CrohnsPred: Applications of Deep Learning for Genetic Risk Prediction of Crohn’s Disease
Crohn’s Disease (CD) is a chronic illness that has seen a dramatic increase in global prevalence and incidence in the last two decades [3]. The development of Crohn’s has been partially attributed, amongst other factors, to certain mutations in one’s genes [1]. Most current techniques in Bioinformatics focus on investigating how relevant single genetic mutations are in the development of CD and can fall short in considering important interactions between sparse mutations, which are believed to greatly influence the development of complex illnesses like Crohn’s [2].
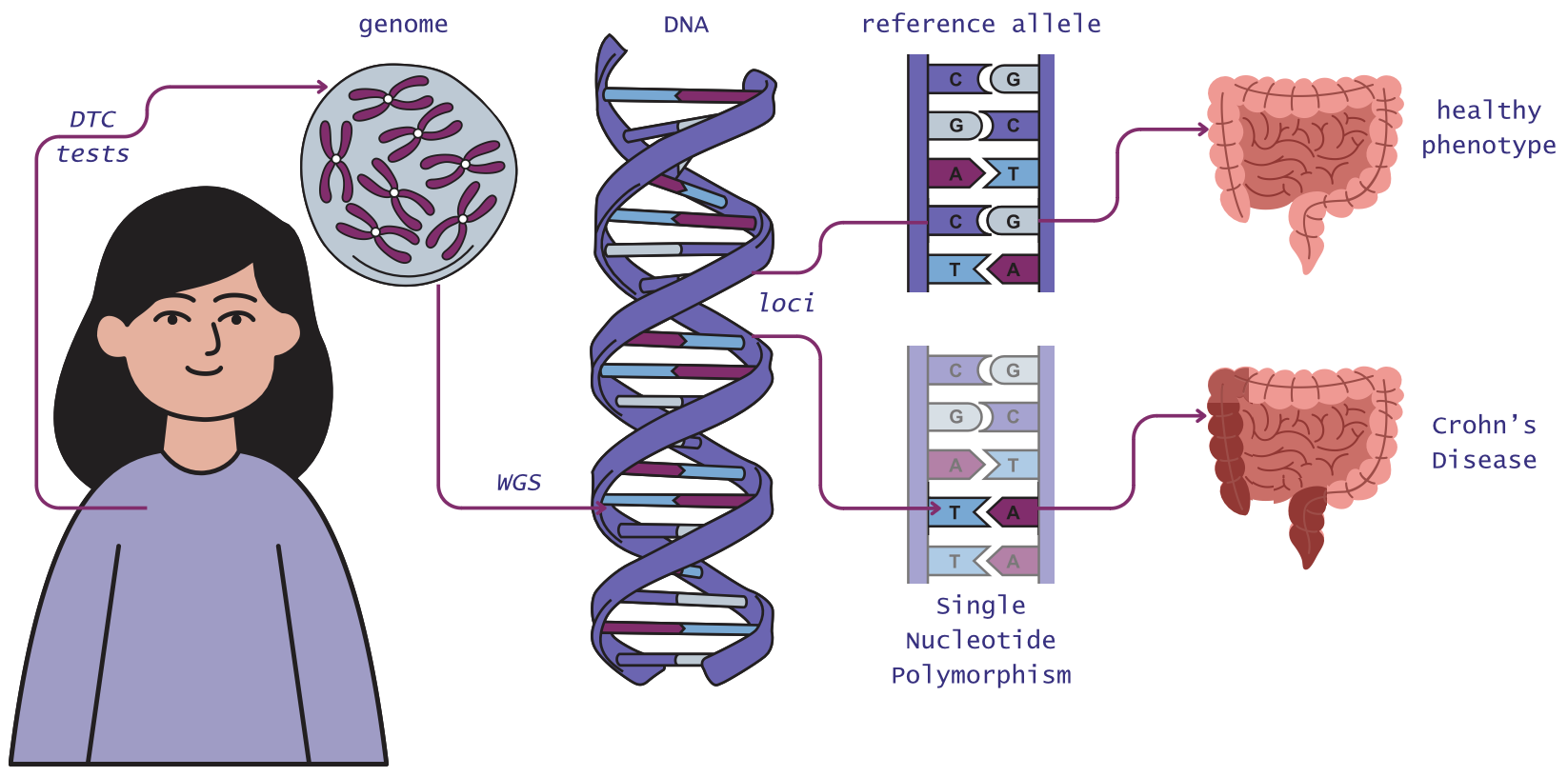
This project proposes a Deep Neural Network, CrohnsPred, for the prediction of an individual’s Genetic Risk of Crohn’s Disease based on sample-level genotype mutation data. This approach leverages the ability of DNNs to identify patterns and discover links amongst great amounts of data, which have been shown to outperform various statistical models currently used for the polygenic score calculation for other polygenic diseases [5]. To preserve data anonymity and reduce ethical concerns, the genetic and phenotypic data used for the Network training and testing was created as synthetic data through the HAPNEST software [4].
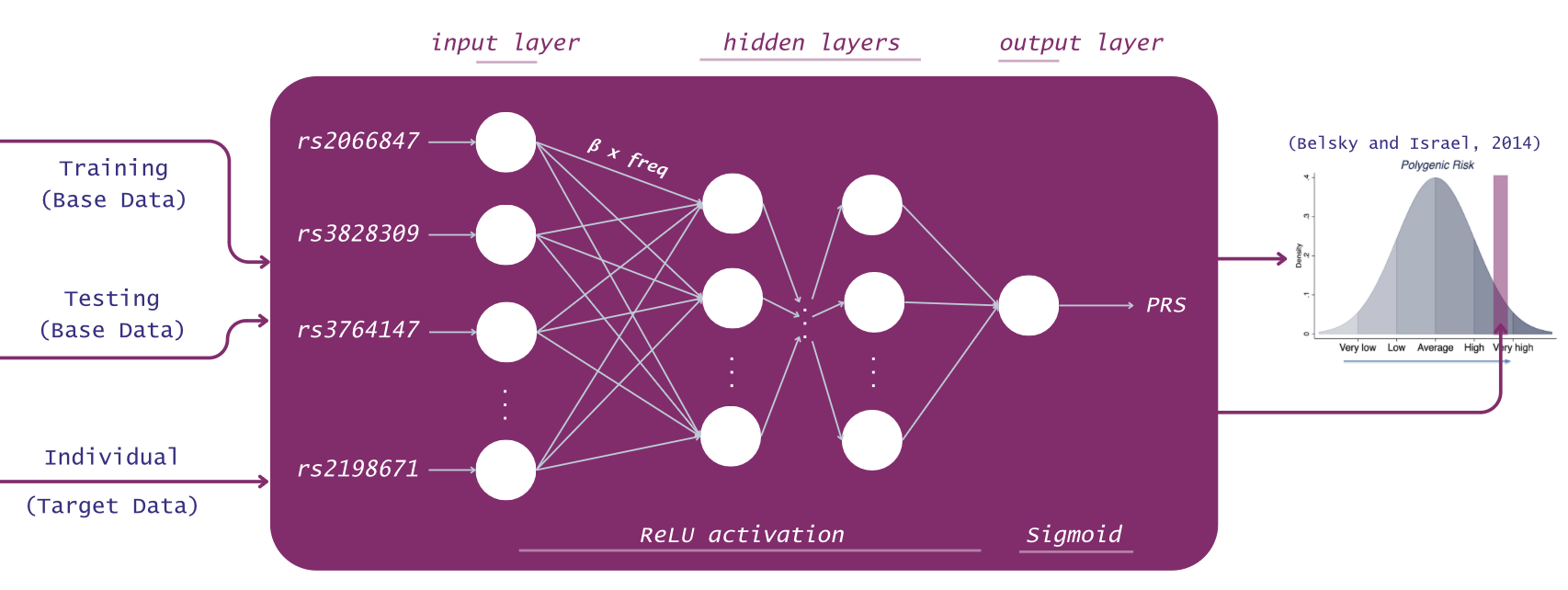
The prediction tool is integrated into a publicly accessible web-based dashboard, which allows the users to get a personalized prediction and access their score through eye-catching graphics and visualizations for a facile understanding of the results.
Implemetation and Data Analysis
Webapp interface
References
[1] Aleksejs Sazonovs, Stevens, C., Guhan Ram Venkataraman, Yuan, K., Avila, B.E., Abreu, M.T., Ahmad, T., Matthieu Allez, Atzmon, G., Baras, A., Jc, B., Nir Barzilai, Laurent Beaugerie, Beecham, A., Bernstein, Ç.N., Bitton, A., Bernd Bokemeyer, Chan, A., Chung, D.C. and Cleynen, I. (2021). Sequencing of over 100,000 individuals identifies multiple genes and rare variants associated with Crohn’s disease susceptibility. medRxiv (Cold Spring Harbor Laboratory). doi:https://doi.org/10.1101/2021.06.15.21258641.
[2] Behravan, H., Hartikainen, J.M., Tengström, M., Pylkäs, K., Winqvist, R., Kosma, V., Mannermaa, A., 2018. Machine learning identifies interacting genetic variants contributing to breast cancer risk: A case study in Finnish cases and controls. Sci. Rep. 8 , 13149. https://doi.org/10.1038/s41598-018-31573-5.
[3] Roda, G., Chien Ng, S., Kotze, P.G., Argollo, M., Panaccione, R., Spinelli, A., Kaser, A., Peyrin-Biroulet, L. and Danese, S. (2020). Crohn’s disease. Nature Reviews Disease Primers, [online] 6(1), pp.1–19. doi:https://doi.org/10.1038/s41572-020-0156-2.
[4] Wharrie, S., Yang, Z., Raj, V., Monti, R., Gupta, R., Wang, Y., Martin, A., O’Connor, L.J., Kaski, S., Pekka Marttinen, Pier Francesco Palamara, Lippert, C. and Ganna, A. (2022). HAPNEST: efficient, large-scale generation and evaluation of synthetic datase ts for genotypes and phenotypes. bioRxiv (Cold Spring Harbor Laboratory). doi:https://doi.org/10.1101/2022.12.22.521552.
[5] Zhou, X., Chen, Y., Ip, F.C.F., Jiang, Y., Cao, H., Lv, G., Zhong, H., Chen, J., Ye, T., Chen, Y., Zhang, Y., Ma, S., Lo, R.M.N., Tong, E.P.S., Mok, V.C.T., Kwok, T.C.Y., Guo, Q., Mok, K.Y., Shoai, M. and Hardy, J. (2023). Deep learning-based polygenic risk analysis for Alzheimer’s disease prediction. Communications Medicine, [online] 3(1), pp.1–20. doi:https://doi.org/10.1038/s43856-023- 00269-x.
